- Simplified experimental scenarios of 3C, 4C, 5C, ChIA-PET and Hi-C technology
- The methodology of importing the interactions into the database
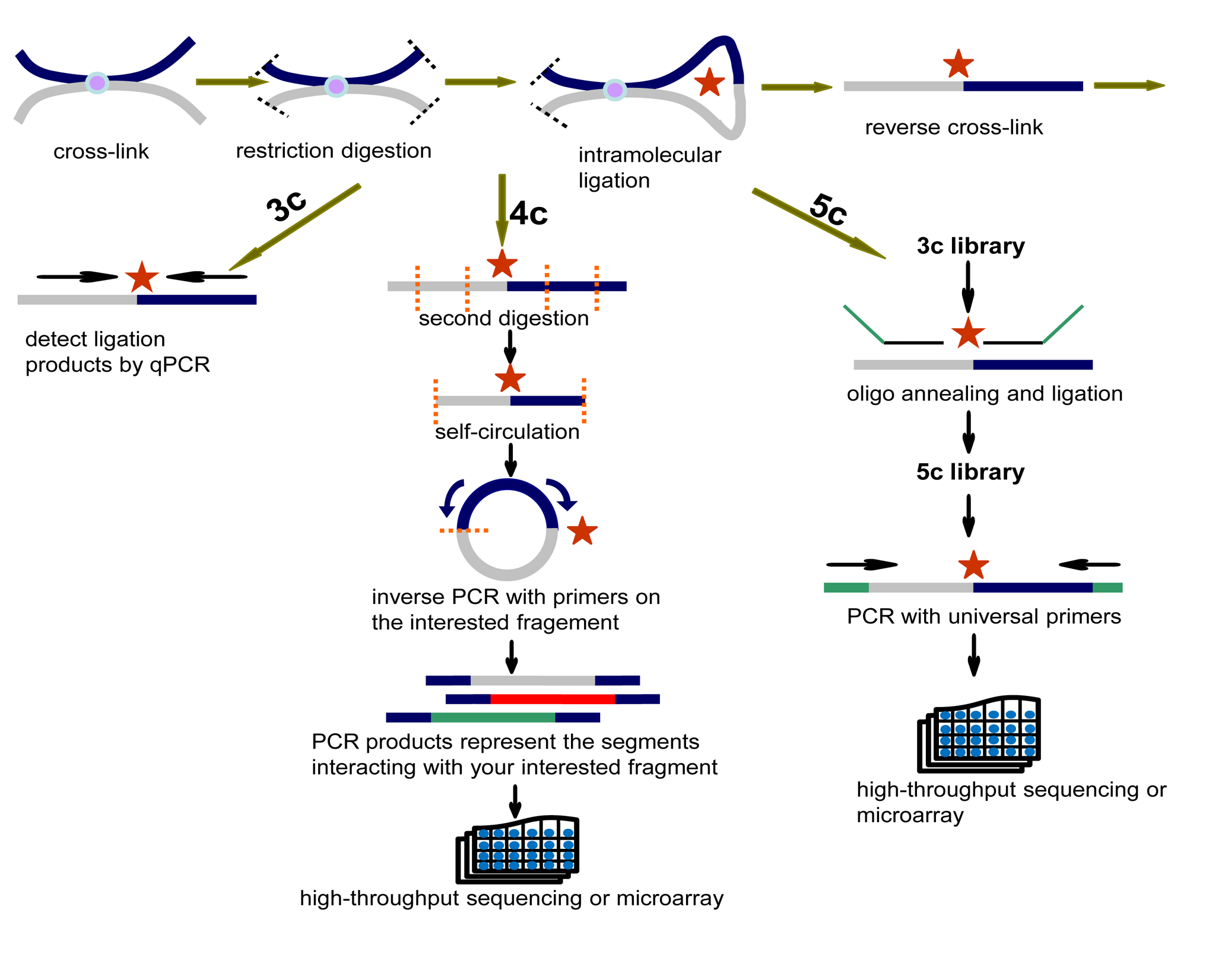
1. Simplified experimental scenarios of 3C, 4C, 5C, ChIA-PET and Hi-C are as follows:
- 3C(Chromosome Conformation Capture): one to one
- 4C(circular 3C or 3C-on-chip): one to all
- 5C(Chromosome Conformation Capture Carbon Copy): many to many
- ChIA-PET(Chromatin Interaction Analysis with Paired-End-Tag): all interactions bound by specific factor
- Hi-C: all to all

2. The methodology of importing the interactions into the database
- We collected 91 sets of chromatin interaction data and then formatted them into "frag_1, frag_2, contact, FDR (false discovery rate) and p-value". Frag_1 and frag_2 were the two interactional fragments, each represented by a genomic region: chromosome: start-end. FDRs/p-values came directly from original literature or were counted according to literature. If counted, FDRs were equal to expected interactions divided by observed interactions. We kept the credible interaction pairs with the standard FDR<0.05 or p<0.01. If FDRs/p-values did not exist for some literature, we kept those datasets by replacing with "NA" on the basis of confidence.
- All interaction data were converted into the current version (hg38 and mm10) by UCSC liftOver and then were annotated with genes, enhancers and SNPs by analysis pipeline (deposited in github: https://github.com/xiexiaowei/CCSI), which was based on bedtools (intersectBed). Eventually, the data format was transformed into two interactional fragments with promoters, enhancers and SNPs, along with contact, FDR and p-value for them.
- Importing annotated interaction data to mySQL database with python scripts named to_mysql.py (deposited in github: https://github.com/xiexiaowei/CCSI).
- Based on PHP scripts, thus users are able to search chromatin interactions in the CCSI website.
